Liquid biopsy using mFISH
Existing assays/assays in development for liquid biopsy exploit circulating tumor cells, cell-free nucleic acids, exosomes or protein antigens, but rarely noncancerous cells in peripheral blood, which include granulocytes, peripheral blood mononuclear cells (PBMCs), tumour-educated platelets, circulating endothelial cell clusters derived from the tumour, and potentially other still-to-be-discovered cell types and cell aggregates.
Liquid biopsies are minimally invasive and are faster and cheaper
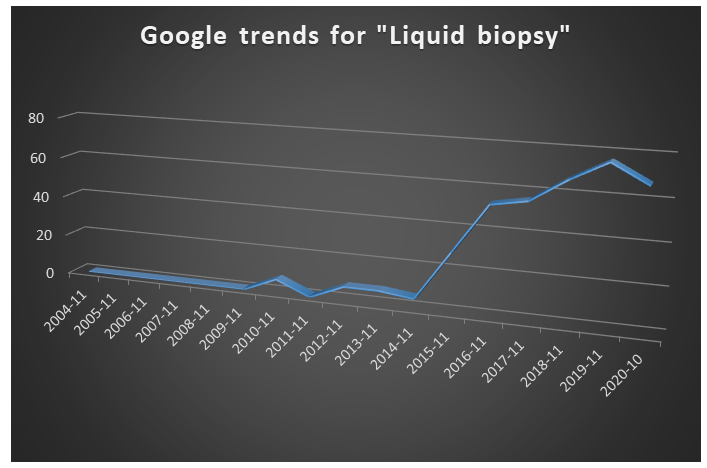
A quick search on google trends for the term ”liquid biopsy” shows an increasing interest since 2015
A typical workflow in liquid biopsy involves collection of biological fluids, performing molecular assays and then data mining to quantify useful and relevant disease biomarkers. Many molecular assays adopt a “grind and find” biochemical approach wherein information from multiple cells are averaged to obtain appropriate clinical information. Typically, levels of 6-8 biomarkers are measured from each investigation.
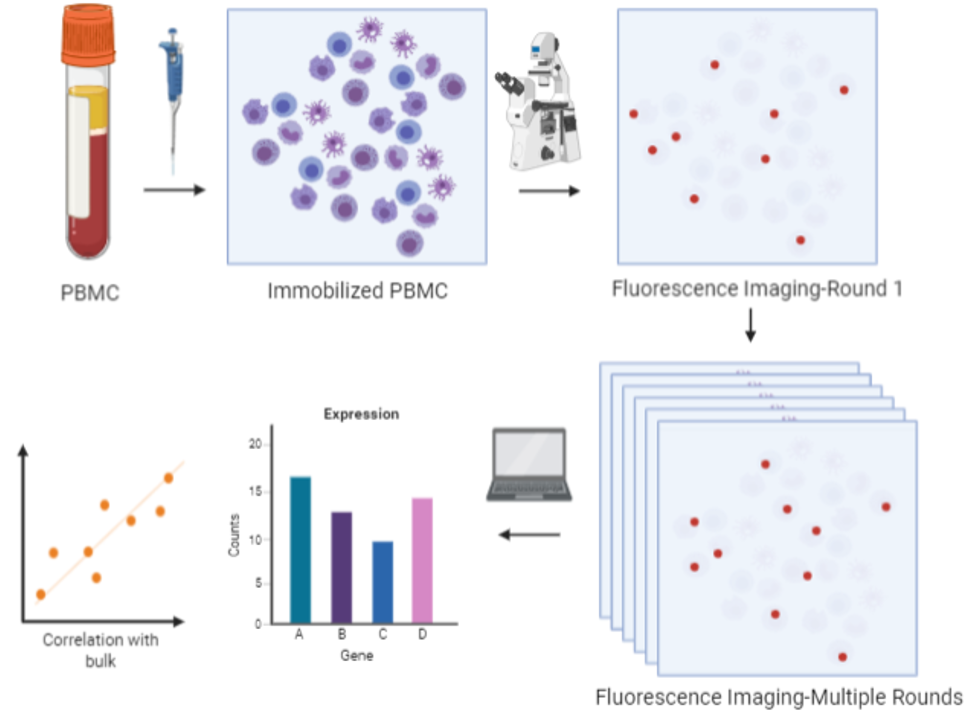
Using our spatial omics technology called mFISH, the cellular spatial information enabling one to obtain correlations between the location of the biomarker and the health status of the individual from whom the sample was collected. mFISH provides a multiplexing advantage wherein >100 biomarkers can be probed at single shot at greater sensitivity than current methods. These advantages potentially provide greater speed, precision and convenience at lower cost for clinical diagnoses.
We primarily plan to use this approach to investigate a novel therapeutic strategy for cancer patients called immunotherapy wherein the patient’s natural defenses are boosted to fight cancer. We will perform mFISH on blood samples to profile potential cancer biomarkers, including those that are known to mediate immune responses, expressing in the peripheral immune cells for cancer diagnosis and longitudinal monitoring.
