Immune checkpoint inhibitors such as anti-CTLA4 and anti-PD1 drugs have revolutionized the field of cancer therapy. However, sustained treatment responses are observed only in a minority of colorectal cancer patients (MSI-H patients), highlighting the need to better understand the functions and interactions of tumor, stromal and immune cells in the colorectal cancer TME.
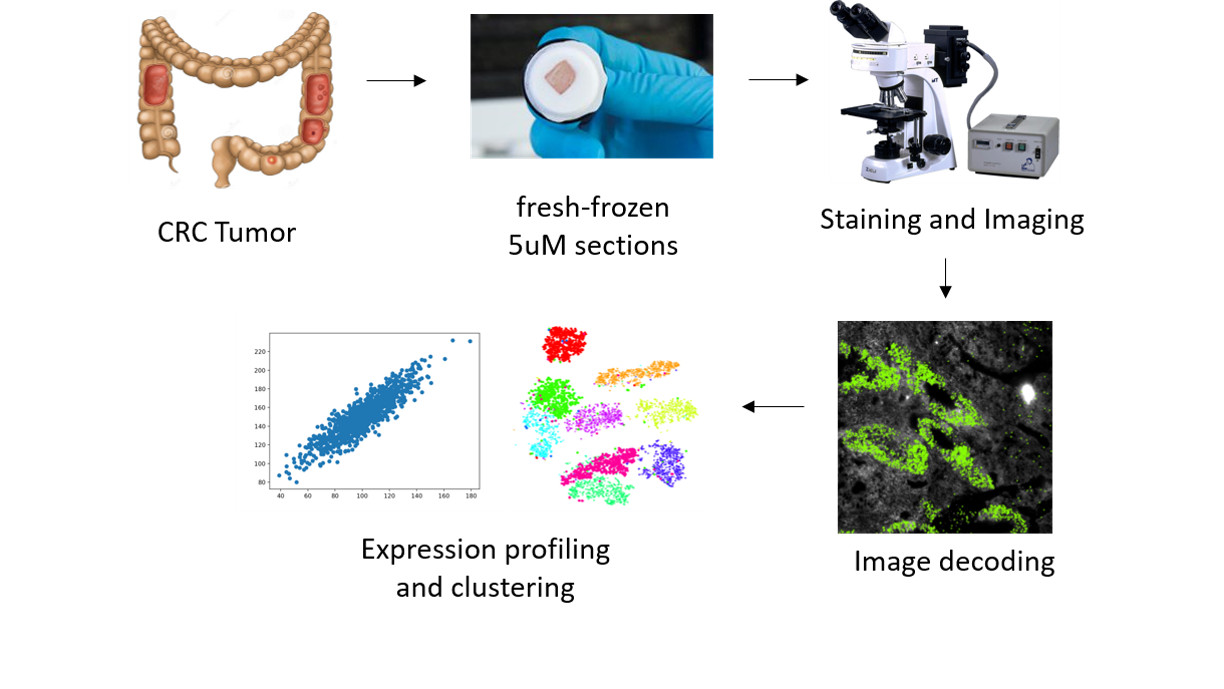
In this project, we aim to use multiplex error robust fluorescence-in-situ-hybridization (mFISH) to create accurate, sub-cellular-resolution cellular profiles of the colorectal TME and study the roles and interactions of different cell types. We will also compare the immune alterations between responders and non-responders to immunotherapy, and identify (spatial) biomarkers that may shed light on the mechanisms of immunotherapy responses.
Compared with existing methods for characterizing tumor cell types that rely on sequencing information alone, our mFISH approach stands out in its ability to provide the much-desired in situ spatial information, thus enhancing the accuracy in cell type identification. Such information is key for understanding the molecular and cellular interactions within the tumor microenvironment.